Playing with RStudio
I've been trying out RStudio [1] for plotting - eg the candlestick plots earlier in the thread were produced with that. Let's see how that would work if I wanted to plot some shift curves from rpm vs speed data.
First, I'll use my Python script to extract the data as CSV's. The script is fairly simple, asammdf API [2] does the heavy lifting, my script just basically takes the command line parameters and based on those tells asammdf what to do. Here, we feed two MDF files in, extract signals using dbc
filters file, filter down to three specific signals, resample to 100Hz and output CSV:
Code:
$ python3 mdf.py 00000001-649487D1.MF4 00000002-64948975.MF4 --dbc curated.dbc --channel CAN2.engine_0x280.rpm --channel CAN1.mfd_0x320.speed_actual --channel CAN1.mfd_0x320.accelerator_on --resample 0.01 --to_file data.csv --to_format csv
ldf is not supported
xls is not supported
xlsx is not supported
from : -1
to : -1
resample : 0.01
dbc : ['curated.dbc']
files : ['00000001-649487D1.MF4', '00000002-64948975.MF4']
channels : ['CAN2.engine_0x280.rpm', 'CAN1.mfd_0x320.speed_actual', 'CAN1.mfd_0x320.accelerator_on']
reading files...
applying dbc files...
filtering channels...
resampling...
exporting CSV...
Channels:
CAN2.engine_0x280.rpm
unit : U/min
timestamps : 232591
first : 0.0
last : 2325.8999999999505
samples : 232591
first : 819.5
last : 830.5
min : 793.25
max : 3189.917085424477
CAN1.mfd_0x320.speed_actual
unit : km/h
timestamps : 232591
first : 0.0
last : 2325.8999999999505
samples : 232591
first : 0.00526
last : 0.00526
min : 0.00526
max : 130.14661660747612
CAN1.mfd_0x320.accelerator_on
unit :
timestamps : 232591
first : 0.0
last : 2325.8999999999505
samples : 232591
first : 0
last : 0
min : 0
max : 1
As an output, I get summary of each channel with some quick stats like number of samples, range of timestamps and range of samples. These just help to estimate dataset size and quickly see if eg there's any outrageously high sample values. Due to resampling we have exactly same amount of samples in all channels and their timestamps all start from 0. In this example we got about 230k samples over 2300 seconds on all three channels so resampling to 100Hz seemed to work as intended.
Looking at the CSV files produced, I got just two files although I had three channels. This is because speed_actual and accelerator_on signals were from same channel group - asammdf exports by groups:
Code:
$ ls -lanh
total 228432
drwxr-xr-x@ 8 501 20 256B Jun 25 10:42 .
drwxr-xr-x 28 501 20 896B Jun 24 18:24 ..
-rw-r--r-- 1 501 20 50M Jun 22 20:41 00000001-649487D1.MF4
-rw-r--r-- 1 501 20 44M Jun 22 20:48 00000002-64948975.MF4
-rw-r--r-- 1 501 20 13K Jun 24 18:35 curated.dbc
-rw-r--r-- 1 501 20 7.1M Jun 25 10:29 data.ChannelGroup_0_CAN2_-_message_engine_0x280_0x280.csv
-rw-r--r-- 1 501 20 8.9M Jun 25 10:29 data.ChannelGroup_1_CAN1_-_message_mfd_0x320_0x320.csv
-rw-r--r-- 1 501 20 6.5K Jun 25 10:08 mdf.py
The contents of CSV will look like this:
Code:
$ head data.ChannelGroup_1_CAN1_-_message_mfd_0x320_0x320.csv
timestamps,CAN1.mfd_0x320.speed_actual,CAN1.mfd_0x320.accelerator_on
0.0,0.005260000005364418,0
0.009999999999999787,0.005260000005364418,0
0.019999999999999574,0.005260000005364418,0
0.02999999999999936,0.005260000005364418,0
0.03999999999999915,0.005260000005364418,0
0.049999999999998934,0.005260000005364418,0
0.05999999999999872,0.005260000005364418,0
0.06999999999999851,0.005260000005364418,0
0.0799999999999983,0.005260000005364418,0
RStudio, or R statistical language in general, is pretty nice for working with data and plotting as you describe both the data manipulations and graphs as text commands on a command prompt. This allows, for example, graph "recipes" to be simply copied and pasted, yet repeatable graphs can be produced anywhere. I'm a complete noobie on these and basically just learning stuff on the go in the search of optimum workflow.
So, let's first load the CSV files in to R. To do this, we first need to install and load the libraries we're going to use: readr [3] for CSV reading and ggplot2 [4] for plotting. Installing the libraries needs only to be done on the first go, later on it's enough to just library() command to load them. As the CSV export produced rather elaborate column names let's also rename the columns for easier use:
Code:
> install.packages(readr)
> install.packages(ggplot2)
> library(readr)
> library(ggplot2)
>
> rpm <- read_csv("/<FULL PATH>/data.ChannelGroup_0_CAN2_-_message_engine_0x280_0x280.csv")
Rows: 232591 Columns: 2
── Column specification ──────────────────────────────────────────────────────────────────────
Delimiter: ","
dbl (2): timestamps, CAN2.engine_0x280.rpm
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
>
> speed <- read_csv("/<FULL PATH>/data.ChannelGroup_1_CAN1_-_message_mfd_0x320_0x320.csv")
Rows: 232591 Columns: 3
── Column specification ──────────────────────────────────────────────────────────────────────
Delimiter: ","
dbl (3): timestamps, CAN1.mfd_0x320.speed_actual, CAN1.mfd_0x320.accelerator_on
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
>
> colnames(rpm)[2] = "rpm"
> colnames(speed)[2] = "speed"
> colnames(speed)[3] = "accelerator_on"
After having the datasets loaded, I can combine these to a single table joining by the timestamp. This is strictly not necessary, separate datasets can be used as well but it's kind of easier to work with all data in a same table. Just entering the table name will show some rows so we can see what the data looks like:
Code:
> my_data <- merge(rpm, speed, by='timestamps')
> my_data
timestamps rpm speed accelerator_on
1 0.00 819.5000 0.00526 0
2 0.01 815.0000 0.00526 0
3 0.02 815.0000 0.00526 0
...
248 2.47 827.7500 0.00526 0
249 2.48 822.8500 0.00526 0
250 2.49 822.7500 0.00526 0
[ reached 'max' / getOption("max.print") -- omitted 232341 rows ]
Now, since I have coasting I know my graph will be little bit messed up if I plot the coasting parts. That's why I picked the accelerator_on column - we can now filter away sample rows where the accelerator was 0 (not pressed):
Code:
> my_data2 <- my_data[my_data$accelerator_on > 0,]
> my_data2
timestamps rpm speed accelerator_on
5001 50.00 1664.188 22.05591 1
5002 50.01 1669.250 22.10432 1
...
5248 52.47 1600.500 34.05493 1
5249 52.48 1603.370 34.11644 1
5250 52.49 1610.750 34.17795 1
[ reached 'max' / getOption("max.print") -- omitted 110473 rows ]
The new dataset has 110k rows remaining, there was quite a bit of coasting or idling in this data set.
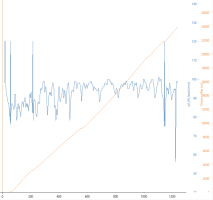
Finally, we have the data prepared and I can generate a plot. In this example, I'm giving a title to the graph, instructing on how I want my x and y axis to be named and what limits and breaks to use on them. Then, I overlay a scatter plot (geom_point) and tell x axis should represent the speed whily y should be rpm.
Code:
> ggplot() +
+ ggtitle("RPM vs speed") +
+ scale_y_continuous(name = "RPM", limits = c(0, 4000), breaks = seq(0, 5000, 1000)) +
+ scale_x_continuous(name = "speed", limits = c(0, 140), breaks = seq(0, 140, 20)) +
+ geom_point(data = my_data2, mapping = aes(x = speed, y = rpm), color = 'darkcyan')
As a result, I will get a plot like below. The gears can be pretty well seen except the 1st is little bit mushy. I have DSG TVS mapping, would be interesting to compare a similar plot for non-TVS -mapped box


While RStudio and ggplot2 are really nice, they don't quite shine in plotting multiple signals in the same graph. In data science world, which these tools are really created for, multiple axis is considered confusing [5]. So, there's no easy way to add eg couple of digital signals on top / under a plot, separate plots must be created. That's quite a shame and probably means the search for ultimate plotting tool continues - but definitely RStudio is a valuable addition to the arsenal.

[1]
Posit
[2]
API — asammdf "7.3.12" documentation
[3]
GitHub - tidyverse/readr: Read flat files (csv, tsv, fwf) into R
[4]
GitHub - tidyverse/ggplot2: An implementation of the Grammar of Graphics in R
[5]
Why not to use two axes, and what to use instead